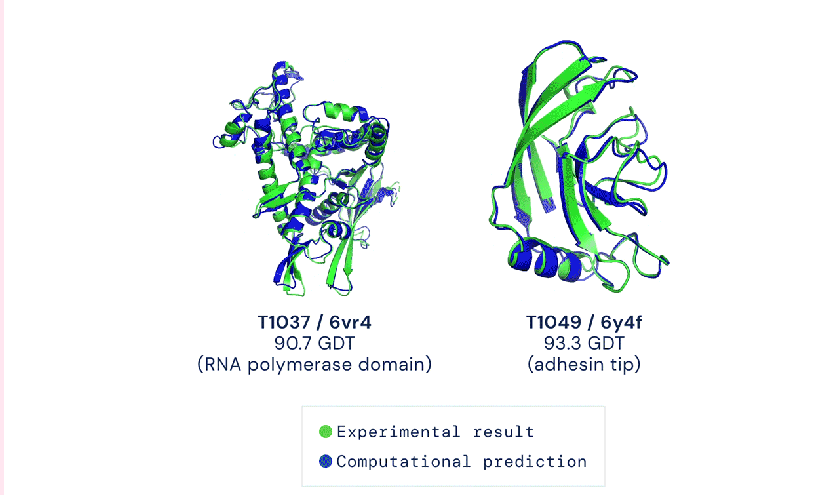
Two examples of protein targets in the free modelling category show AlphaFold’s prediction compared to the shape of proteins determined by experimental results. AlphaFold’s predictions are in blue and the experimental results are in green. Screenshot from DeepMind.
DeepMind, the Google subsidiary that has been beating chess and Go players with artificial intelligence, has set its sights on cracking a decades-old problem: predicting the structures of proteins.
At a biennial challenge where participants must blindly predict the structure of 100 proteins based on their amino acid sequences, a system developed by DeepMind captured researchers’ attention when it predicted their shape with a high level of accuracy.
Called AlphaFold, the system determined the shape of around two-thirds of the proteins with an accuracy comparable to time-consuming laboratory experiments. Its accuracy with most of the other proteins was also high, according to results shared by CASP (the Community Wide Experiment on the Critical Assessment of Techniques for Protein Structure Prediction) on Monday. The results were compared to the shape of proteins discovered in the lab and were assessed by independent scientists.
This is an important breakthrough because the shape of proteins is closely linked with their function, but it is difficult to predict a protein’s structure based on its amino acid sequence. Proteins can theoretically fold into a multitude of shapes before setting into their final structure. It can take years of research, and expensive equipment, to work out their shape.
“Proteins are extremely complicated molecules, and their precise three-dimensional structure is key to the many roles they perform, for example the insulin that regulates sugar levels in our blood and the antibodies that help us fight infections. Even tiny rearrangements of these vital molecules can have catastrophic effects on our health, so one of the most efficient ways to understand disease and find new treatments is to study the proteins involved,” John Moult, a computational biologist at the University of Maryland in College Park who co-founded CASP, said in a news release.

A Deep-dive Into Specialty Pharma
A specialty drug is a class of prescription medications used to treat complex, chronic or rare medical conditions. Although this classification was originally intended to define the treatment of rare, also termed “orphan” diseases, affecting fewer than 200,000 people in the US, more recently, specialty drugs have emerged as the cornerstone of treatment for chronic and complex diseases such as cancer, autoimmune conditions, diabetes, hepatitis C, and HIV/AIDS.
London-based DeepMind has been working on AlphaFold for four years. It also beat the other teams in the last CASP challenge in 2018, but did so by a much larger margin in the most recent year.
The model’s accuracy is measured using the Global Distance Test, which approximately measures the percentage of amino acid residues within a certain distance from the correct position. In a scale of 1 to 100, DeepMind’s latest AlphaFold system scored a median of 92.4 across all targets.
For the latest iteration of AlphaFold, DeepMind designed a neural network that interprets a protein’s structure as a “spatial graph.” It trained the system on 170,000 protein structures from the protein data bank as well as databases with proteins whose structure was unknown.
This allowed the system to determine structures in a matter of days, the team who developed it wrote in a blog post. An internal confidence measure also indicated which parts of each predicted protein structure are reliable.
What does this all mean? It could have broad implications for drug discovery and better understanding specific diseases. Andrei Lupas, director of the Max Planck Institute for Developmental Biology and a CASP assessor, stated that the system helped his team solve a protein structure that they were stuck on for close to a decade.
Andriy Kryshtafovych, a researcher at UC Davis and one of the judges, described the result as a “triumph for team science,” crediting the collaborative work of researchers over the years to reaching this achievement.
“Being able to investigate the shape of proteins quickly and accurately has the potential to revolutionize life sciences,” he said in a news release. “Now that the problem has been largely solved for single proteins, the way is open for development of new methods for determining the shape of protein complexes – collections of proteins that work together to form much of the machinery of life, and for other applications.”